前言
计算单点突变自由能是分子稳定性改造的”老套路”。FoldX以及ddg_monomer application已经久经实战考验(具体用法参考前人总结:http://bioengx.hk1.91site.net/rosetta-ddg_monomer/)。今天我们就来说说Rosetta新一代的预测方法: Cartesian_ddG。
1 Cartesian_ddG方法概述
参考: Hahnbeom Park, Philip Bradley, Per Greisen Jr., Yuan Liu, Vikram Khipple Mulligan, David E Kim, David Baker, and Frank DiMaio (2016) “Simultaneous optimization of biomolecular energy function on features from small molecules and macromolecules”, JCTC.
Cartesian_ddG是一种新的采样方法,与ddg_monomer不同,对于骨架柔性的计算并不需要大量重复的强限制约束,而是采取了卡迪尔空间的优化来允许小幅度的骨架运动。
在计算∆∆G时,Cartesian_ddG方法使用两步Relax的方法:
-预先使用FastRelax进行优化,确定野生型以及点突变氨基酸最佳的侧链排布方式
-再使用卡迪尔空间版本的FastRelax对局部区域(点突变位置6埃范围内)进行彻底放松。
-评估野生型以及突变型蛋白的能量差,并使用能量函数特异性的校正因子对∆∆G值进一步矫正,以对应实验测定值单位kcal/mole。
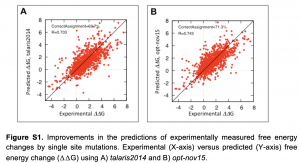
效果:更快、更准。
Cartesian_ddG(使用opt-nov15函数)比ddg_monomer最佳Protocol(row16)要快上10倍有余。并且Cartesian_ddG预测∆∆G与实验∆∆G的皮尔森相关性从0.703升至0.743。
2 Cartesian_ddG的使用方法
Cartesian_ddG的使用方法十分简单。
-预优化野生型蛋白结构
-准备mutfile文件
-运行Cartesian_ddG
-分析∆∆G值
2.1 预优化野生型蛋白结构
创建relax_flag文件,文件内容如下:
-use_input_sc
-constrain_relax_to_start_coords
-ignore_unrecognized_res
-nstruct 20
-relax:coord_constrain_sidechains
-relax:cartesian-score:weights ref2015_cart # 务必设为ref2015_cart
-relax:min_type lbfgs_armijo_nonmonotone # 能量最小化的算法
-relax:script cart2.script
创建cart2.script文件,文件内容如下:(无需额外更改参数)
switch:cartesian
repeat 2
ramp_repack_min 0.02 0.01 1.0 50
ramp_repack_min 0.250 0.01 0.5 50
ramp_repack_min 0.550 0.01 0.0 100
ramp_repack_min 1 0.00001 0.0 200
accept_to_best
endrepeat
进行relax优化,打开终端并输入:
relax.mpi.macosclangrelease -s $pdb @relax_flag
打开relax的打分文件,查看最低能量的结构pdb编号,并以此作为出发的输入结构。
2.2 准备mutfile文件
单点突变的mutfile文件的格式如下:
注意!此处的89为pose序号,而非PDB中的残基号,设置错误一定会报错。
total 1 # mutfile中总突变数量
1 # 本轮突变氨基酸个数,单点突变设置为1.
T 89 A # 将89位的T突变为A
将文件保存为T89A.mutfile
2.3 运行Cartesian_ddG
Cartesian_ddG有两种运行模式: 蛋白稳定性预测模式(Protein stability mode)和 相互作用界面模式(Interface mode)。
1)蛋白稳定性预测模式(Protein stability mode)
创建cartddg_flag文件, 内容如下:
-ddg:iterations 5 # 默认为3,可以根据自身计算资源调整。
-ddg::cartesian
-ddg::dump_pdbs False # 是否输出突变后的蛋白PDB文件,推荐False,否则文件夹会很乱
-bbnbrs 1 # 骨架自由度,额外考虑线性邻居氨基酸的数目,1代表 邻近的3个氨基酸骨架自由度被考虑在计算中。
-fa_max_dis 9.0 # 控制范德华和溶剂化能量计算的范围,默认为6埃以内的氨基酸被考虑。
-score:weights ref2015_cart # 务必设为ref2015_cart
运行ddg计算,打开终端并输入,这么就会开始计算T89A的∆∆G。
cartesian_ddg.linuxgccrelease -s [inputpdb] @cartddg_flag -ddg:mut_file T89A.mutfile
2 )相互作用界面模式(Interface mode)
该模式开发者并未做详细测试,斟酌使用,此处笔者未做测试。如有需要请参考: https://www.rosettacommons.org/docs/latest/cartesian-ddG
3 结果分析
运行完毕后,文件夹中会产生T89A.ddg这个文件。打开后,文件内容如下:
COMPLEX: Round1: WT: -333.782 fa_atr: -840.140 fa_rep: ………….
COMPLEX: Round2: WT: -333.782 fa_atr: -840.140 fa_rep: ………….
COMPLEX: Round3: WT: -333.782 fa_atr: -840.140 fa_rep: ………….
COMPLEX: Round4: WT: -333.782 fa_atr: -840.140 fa_rep: ………….
COMPLEX: Round5: WT: -333.782 fa_atr: -840.140 fa_rep: ………….
COMPLEX: Round1: MUT_89ALA: -330.352 fa_atr: -834.516 ………….
COMPLEX: Round2: MUT_89ALA: -330.352 fa_atr: -834.516 ………….
COMPLEX: Round3: MUT_89ALA: -330.352 fa_atr: -834.516 ………….
COMPLEX: Round4: MUT_89ALA: -330.352 fa_atr: -834.516 ………….
COMPLEX: Round5: MUT_89ALA: -330.352 fa_atr: -834.516 ………….
此处Round代表第几次迭代,根据-ddg:iterations设置定义。
WT:代表本轮能量计算,野生型的能量为-333.782。
MUT_89ALA:代表本轮能量计算,突变型的能量为-330.352。
我们需要将WT和MUT_89ALA平均后,用MUT能量平均值减去WT能量平均值就可以得到T89A的∆∆G值,为3.43。
稳定性变化评判标准:(务必根据自身需要调整)
∆∆G < -1.0 代表点突变为蛋白带来稳定效应; ∆∆G > 1.0 代表点突变为蛋白带来不稳定效应;
备注: 如果需要批量分析和点突变扫描,请使用脚本生成多个mutfile,然后按照以上流程进行计算。








purchase generic Accutane online discreetly: cheap Accutane – cheap Accutane
tadalafil online no rx [url=http://tadalafilfromindia.com/#]tadalafil online no rx[/url] generic Cialis from India
generic Cialis from India: tadalafil 2.5 mg cost – tadalafil online no rx
Zoloft online pharmacy USA: Zoloft online pharmacy USA – Zoloft Company
lexapro price comparison [url=https://lexapro.pro/#]Lexapro for depression online[/url] lexapro 15mg
order propecia tablets: order propecia without rx – cheap Propecia Canada
It’s very effortless to find out any matter on net as compared to textbooks, as I found this article at this web site.
My web page; Best Seo
Thank you, I’ve recently been searching for info approximately this subject for ages and yours is
the greatest I’ve discovered so far. But, what concerning the bottom line?
Are you certain about the supply?
my website … Remodelling
Good day I am so happy I found your website, I really found you by
mistake, while I was looking on Google for something else, Anyways
I am here now and would just like to say thank you for a remarkable post and a all
round entertaining blog (I also love the theme/design), I don’t
have time to read through it all at the minute but I have book-marked it and also added
your RSS feeds, so when I have time I will be back to read more, Please do
keep up the fantastic jo.
Here is my web-site … roofing contractor near me
https://zoloft.company/# cheap Zoloft
Hi, I desire to subscribe for this blog to take most recent updates,
thus where can i do it please help out.
Here is my blog post :: hydro jetting services
https://zoloft.company/# sertraline online
http://tadalafilfromindia.com/# Tadalafil From India
Thank you for sharing your thoughts. I really appreciate your efforts and I am waiting for your further write ups thanks
once again.
My site :: roofing companies
Wonderful beat ! I wish to apprentice while you amend your website,
how can i subscribe for a blog site? The account helped
me a acceptable deal. I had been tiny bit acquainted of this your broadcast provided bright clear
concept
my web blog; roof repair services Shelbyville
Because the admin of this web site is working, no uncertainty very soon it will
be well-known, due to its quality contents.
Stop by my site :: banquet halls
Fascinating blog! Is your theme custom made or did you download it from
somewhere? A design like yours with a few simple adjustements would really make my blog stand out.
Please let me know where you got your theme. Thanks
My web blog: site
It’s not my first time to pay a visit this web
site, i am browsing this web site dailly and take nice information from here all the time.
Here is my web-site – LLC
https://lexapro.pro/# Lexapro for depression online
I just could not depart your web site prior to suggesting
that I actually enjoyed the standard information a person provide on your
guests? Is going to be again continuously to check up on new
posts
my blog – website
lexapro tablets australia: Lexapro for depression online – lexapro 20
This article offers clear idea in support of the new users of blogging,
that actually how to do blogging and site-building.
Also visit my webpage The Doan Law Firm Personal Injury Lawyers nearby
generic Cialis from India: tadalafil online no rx – cheap Cialis Canada
tadalafil online no rx [url=https://tadalafilfromindia.com/#]generic Cialis from India[/url] tadalafil pills 20mg
https://finasteridefromcanada.com/# cheap Propecia Canada
Hi there, I enjoy reading through your article
post. I like to write a little comment to support you.
My website pool resurfacing
https://lexapro.pro/# buy lexapro online without prescription
I believe this is one of the so much important info for me.
And i’m glad studying your article. However should remark on some
general issues, The site taste is great, the articles is
actually nice : D. Good process, cheers
my blog Military Divorce lawyer near me
https://zoloft.company/# buy Zoloft online without prescription USA
I know this web page offers quality depending posts and extra information,
is there any other web site which gives these things in quality?
my web-site: deck builder services near me
Spot on with this write-up, I seriously think this website needs a lot more attention. I’ll probably be back again to see more, thanks
for the advice!
Also visit my blog post Buy Anonymous Proxy
cheap Cialis Canada: Tadalafil From India – Tadalafil From India
Finasteride From Canada: Finasteride From Canada – cost propecia price
Finasteride From Canada [url=https://finasteridefromcanada.shop/#]Finasteride From Canada[/url] Finasteride From Canada
http://finasteridefromcanada.com/# order cheap propecia no prescription
Simply want to say your article is as amazing. The clarity in your
post is simply nice and that i can think you are knowledgeable on this subject.
Fine together with your permission let me to seize your feed to keep up to date with impending post.
Thanks a million and please continue the enjoyable work.
Here is my web page; shower remodeling
It’s very easy to find out any topic on net as compared to books, as I found
this paragraph at this web page.
My web page :: deck builders
It’s remarkable to pay a quick visit this website and reading the
views of all mates regarding this paragraph, while
I am also keen of getting know-how.
Here is my homepage; Bedrock water heater installation near me
buy Cialis online cheap: buy Cialis online cheap – tadalafil 5 mg tablet coupon
It’s going to be end of mine day, except before finish I am reading this enormous piece of writing to improve my know-how.
Here is my website … roofing company Centerton AR
buy Cialis online cheap: buy Cialis online cheap – tadalafil online no rx
http://isotretinoinfromcanada.com/# order isotretinoin from Canada to US
https://lexapro.pro/# Lexapro for depression online
Zoloft Company [url=https://zoloft.company/#]cheap Zoloft[/url] Zoloft Company
gabapentin mexican pharmacy [url=https://medimexicorx.com/#]cheap cialis mexico[/url] rybelsus from mexican pharmacy
low cost mexico pharmacy online: legit mexico pharmacy shipping to USA – trusted mexico pharmacy with US shipping
Do you have a spam issue on this website; I also am a blogger, and I was wondering your situation;
we have developed some nice practices and we are looking
to trade strategies with others, why not shoot me an e-mail if interested.
Also visit my web site best shower remodeling near me
http://medimexicorx.com/# MediMexicoRx
This paragraph is actually a good one it assists new net viewers,
who are wishing for blogging.
Here is my website :: roof repair Centerton
https://medimexicorx.shop/# pharmacies in mexico that ship to usa
Great website you have here but I was wondering
if you knew of any user discussion forums that cover the same
topics talked about here? I’d really like to be a part of community where I can get advice from other experienced
people that share the same interest. If you have any recommendations, please let me know.
Thanks!
Feel free to surf to my blog post: Powell’s Plumbing trusted ac repair
bupropion sr online pharmacy [url=https://expresscarerx.online/#]pharmacy magazine warfarin[/url] online pharmacy no presc uk
rybelsus from mexican pharmacy: online mexico pharmacy USA – MediMexicoRx
MediMexicoRx: amoxicillin mexico online pharmacy – mexican pharmacy for americans
https://indiamedshub.com/# legitimate online pharmacies india
how much is viagra at the pharmacy [url=https://expresscarerx.online/#]ExpressCareRx[/url] online pharmacy rx
indian pharmacy online [url=https://indiamedshub.com/#]IndiaMedsHub[/url] pharmacy website india
viagra pills from mexico: trusted mexico pharmacy with US shipping – MediMexicoRx
MediMexicoRx: buy cialis from mexico – MediMexicoRx
IndiaMedsHub [url=https://indiamedshub.com/#]buy medicines online in india[/url] online shopping pharmacy india
https://expresscarerx.org/# ExpressCareRx
buy cialis from mexico: order from mexican pharmacy online – legit mexico pharmacy shipping to USA
You can use Viagra if you have herpes.
Review my homepage: penis enlargement exercise programs
best rated online pharmacy: rx health pharmacy – ExpressCareRx
https://indiamedshub.com/# mail order pharmacy india
MediMexicoRx [url=https://medimexicorx.com/#]isotretinoin from mexico[/url] get viagra without prescription from mexico
does pharmacy sell viagra: ExpressCareRx – cialis certified online pharmacy
https://indiamedshub.com/# india online pharmacy
finasteride mexico pharmacy [url=https://medimexicorx.com/#]order kamagra from mexican pharmacy[/url] mexican pharmacy for americans
proscar pharmacy: rite aid pharmacy viagra cost – simvastatin pharmacy prices
About the time US LEC was based, the ’96 Act encouraged
some CLECs to construct business models primarily based on the UNE-P rules, without investing tens of millions
of dollars on community facilities. “The CLEC model that relied closely on use of the (incumbents’) amenities is certainly challenged if not inviable anymore,” mentioned Al Kurtze, director of government business development for Cap Gemini Ernst &
Young’s Telecom, Media and Entertainment division. CLECs, rivals to
the four large incumbent native carriers within the
U.S., have seen a commonly used business model whittled away by the U.S.
Recent months have been powerful for competitive native alternate carriers (CLECs),
as their allies get gobbled up by rivals and the government dismantles
network-sharing rules. MCI Inc., two of the biggest nationwide carriers that sided with CLECs
on policy points, have announced plans to merge with two of the incumbent
carriers, collectively referred to as the regional Bells. Even with the
consolidation among major carriers, Storey still sees a task for CLECs.
Visit my blog – sportsbook provider
https://indiamedshub.shop/# IndiaMedsHub
I must thank you for the efforts you’ve put in penning this site.
I’m hoping to view the same high-grade content by you later on as well.
In truth, your creative writing abilities has motivated me
to get my own, personal website now
Have a look at my web-site … Roof repair near me
Hi, i read your blog from time to time and i own a similar
one and i was just curious if you get a lot of spam comments?
If so how do you reduce it, any plugin or anything you can suggest?
I get so much lately it’s driving me insane so any support is very much appreciated.
my homepage; Mikita door installation
people’s pharmacy ambien: ExpressCareRx – ExpressCareRx
apotheek online nederland: MedicijnPunt – antibiotica kopen zonder recept
Wow, this piece of writing is good, my sister is analyzing these kinds
of things, thus I am going to let know her.
Also visit my web page – water heater installation
http://tryggmed.com/# influensavaksine 2022 apotek
http://tryggmed.com/# pottaske apotek
What’s up to all, how is all, I think every one is getting more from this website,
and your views are nice for new visitors.
My web-site – Roof repair Phoenix AZ
I was wondering if you ever considered changing the structure of your blog?
Its very well written; I love what youve got to say.
But maybe you could a little more in the way of content so people could
connect with it better. Youve got an awful lot of text
for only having 1 or two images. Maybe you could space it out better?
Visit my page roof repair near me
It’s fantastic that you are getting ideas from this piece of writing as well as from our
dialogue made at this place.
Feel free to visit my blog: painters
Good post. I learn something totally new and challenging on blogs I stumbleupon every day.
It will always be exciting to read through content from other writers and use something from other web sites.
Review my web blog: painting contractor
https://zorgpakket.com/# inloggen apotheek
I every time used to study piece of writing in news papers but now as I am a user of net thus from now I am
using net for articles, thanks to web.
Feel free to visit my web-site: interior painting contractors
https://snabbapoteket.shop/# gratis medicin
ordonnance mГ©dicale en ligne: compralgyl sans ordonnance – antibiotique cystite sans ordonnance
Hey just wanted to give you a quick heads up. The text in your
content seem to be running off the screen in Safari.
I’m not sure if this is a format issue or something to do with web browser compatibility but
I thought I’d post to let you know. The style and design look great
though! Hope you get the issue solved soon. Cheers
my blog post; personal injury attorneys near me
We stumbled over here from a different page and thought I might check things
out. I like what I see so now i’m following you. Look forward to looking over your web page repeatedly.
Have a look at my web blog :: Winkler Kurtz LLP – Long Island Lawyers
Hello all, just wanted to say that Carlospin is top notch online casinos.
Checked it, and slots are fun. You get your money fast, and helpful customer service.
For casino fans, definitely try carlospin casino.
The site is user-friendly, and it’s reliable.
You should give it a go!
Appreciate this info
Great blog! Is your theme custom made or did you download it from
somewhere? A theme like yours with a few simple adjustements would really make my blog stand out.
Please let me know where you got your theme. Appreciate it
Also visit my web page – LLC
Woah! I’m really enjoying the template/theme of this site.
It’s simple, yet effective. A lot of times it’s very hard to get
that “perfect balance” between superb usability and visual appearance.
I must say you’ve done a awesome job with this. Also, the blog loads super quick for me on Chrome.
Exceptional Blog!
my web-site; Mikita window installers
No matter if some one searches for his vital thing, therefore he/she desires
to be available that in detail, thus that thing is maintained over here.
Here is my web blog :: teman
I couldn’t resist commenting. Very well written!
Also visit my webpage – Norwalk Mover
Hi there! Do you know if they make any plugins to help with
SEO? I’m trying to get my blog to rank for some targeted
keywords but I’m not seeing very good gains. If you know of any please share.
Cheers!
Feel free to visit my web blog :: Meigel best bathroom remodeling near me
Hey there, You’ve done an incredible job. I will certainly digg it and personally suggest to my friends.
I’m sure they’ll be benefited from this site.
my webpage: Powell’s Plumbing
Does your site have a contact page? I’m having trouble
locating it but, I’d like to shoot you an e-mail.
I’ve got some suggestions for your blog you might be interested in hearing.
Either way, great website and I look forward to seeing it grow over time.
my web page; Buy Best SEO Backlinks
Quality articles or reviews is the important to be a focus for the viewers to pay a visit the web page, that’s what this site is providing.
Stop by my web site – St Louis roof repairs
This website really has all the information I needed concerning this subject
and didn’t know who to ask.
Also visit my web site NYC
Good day! This post couldn’t be written any better!
Reading this post reminds me of my good old room mate!
He always kept talking about this. I will forward
this page to him. Pretty sure he will have a good read. Thanks
for sharing!
My web page water damage restoration
Whats up very cool blog!! Guy .. Excellent ..
Amazing .. I’ll bookmark your blog and take the feeds also?
I am happy to search out numerous useful info here
in the publish, we’d like work out more strategies in this regard, thanks for sharing.
. . . . .
Also visit my webpage; Pet Urgent Care of Enterprise
This is my first time visit at here and i am genuinely pleassant to read all at
single place.
my blog: Bedrock plumbers in St Louis Park
I know this website presents quality dependent articles or reviews and extra data,
is there any other site which offers these kinds of information in quality?
Also visit my page … residential painting service
Thanks for sharing your thoughts. I truly appreciate your efforts and I
will be waiting for your further write ups thank you
once again.
Review my web blog: roofing companies near me
Heya i am for the first time here. I found this board and I find It truly
useful & it helped me out a lot. I hope to
give something back and help others like you helped me.
Feel free to surf to my blog roofing contractor
We’re a group of volunteers and opening a new scheme in our community.
Your website offered us with valuable info to work on. You have done an impressive
job and our whole community will be grateful to you.
my blog post … FixStop at Alafaya – Phone & Computer Repair
My partner and I stumbled over here from a different page and thought I
might check things out. I like what I see so now i am following you.
Look forward to going over your web page again.
My web-site … DSH home builders near me
I love your blog.. very nice colors & theme. Did you make this website yourself or did you hire someone
to do it for you? Plz respond as I’m looking to
design my own blog and would like to know where
u got this from. cheers
Feel free to surf to my web page; slot 4d terbaru
I am truly thankful to the holder of this web page who has
shared this great post at here.
Also visit my homepage :: hot water heater inspection services
Hi! Quick question that’s entirely off topic. Do you know how to make your site mobile friendly?
My weblog looks weird when browsing from my iphone4.
I’m trying to find a template or plugin that might be able to resolve this issue.
If you have any suggestions, please share. Cheers!
Feel free to surf to my web page :: Vedder roofing contractor Mt Dora FL
Do you have a spam issue on this website; I also am a blogger, and I
was curious about your situation; we have created some nice procedures and we are looking to
exchange methods with others, be sure to shoot me an email if interested.
Stop by my blog – BDE kitchen remodeling near me
whoah this weblog is fantastic i really like studying your articles.
Stay up the great work! You understand, many persons are
searching round for this information, you can aid them greatly.
My site; Resurfacing
Excellent article. Keep posting such kind of information on your site.
Im really impressed by your blog.
Hi there, You’ve performed an incredible job.
I’ll certainly digg it and personally suggest to my friends.
I am confident they’ll be benefited from this website.
Also visit my blog post :: Winkler Kurtz local personal attorneys
This is very interesting, You are a very skilled blogger.
I have joined your feed and look forward to seeking more
of your great post. Also, I’ve shared your web site in my social networks!
Visit my web page :: emergency plumber services
Hey there outstanding blog! Does running a blog similar to this take
a large amount of work? I’ve no expertise in programming however I had been hoping
to start my own blog in the near future. Anyway, should you have any suggestions or techniques
for new blog owners please share. I know this is off subject
however I simply wanted to ask. Thanks a lot!
Also visit my site: sewer repair Payette
whoah this weblog is fantastic i like studying your
posts. Keep up the great work! You know, a lot of people are
hunting round for this info, you can aid them greatly.
Also visit my site – homepage
I think what you said made a great deal of sense. However, what about this?
suppose you typed a catchier post title? I mean, I don’t want
to tell you how to run your blog, but what
if you added something that grabbed people’s attention? I mean Rosetta Cartesian_ddG: 更快更准确的蛋白质稳定性预测方法 – BioEngX is
a little vanilla. You could look at Yahoo’s home page
and note how they create news headlines to grab people interested.
You might add a related video or a pic or two to get people interested about
what you’ve got to say. In my opinion, it could bring your blog a
little livelier.
Also visit my website; ac repair Garden City ID
For newest news you have to pay a quick visit world-wide-web and on the web I found this website as
a most excellent web page for newest updates.
Feel free to visit my webpage: Suburban sump pump replacement
Have you ever considered publishing an ebook or guest
authoring on other blogs? I have a blog based upon on the same topics you discuss and would really like to have you share some stories/information. I know my viewers would
enjoy your work. If you’re even remotely interested, feel free to send me an e-mail.
Here is my web blog :: Caldwell hvac contractor
of course like your website however you have to test the spelling on several of your posts.
Many of them are rife with spelling issues and I to
find it very troublesome to tell the reality however I’ll definitely come again again.
my web page :: P.C.
Hi, i believe that i saw you visited my weblog so i came to go back the want?.I am trying to in finding issues to improve my web site!I suppose its
ok to use a few of your ideas!!
My blog post – webpage
Just wish to say your article is as surprising.
The clarity in your post is simply nice and i can assume you’re an expert on this subject.
Fine with your permission allow me to grab your feed to keep updated with forthcoming post.
Thanks a million and please continue the rewarding work.
Feel free to surf to my web blog local building architect near me
whoah this weblog is fantastic i really like studying your posts.
Keep up the good work! You recognize, a lot of
individuals are searching round for this information, you can help them greatly.
my homepage; architectural lighting near me
I don’t even know how I ended up here, but I thought this post was great. Cheers!
Cheers!
I don’t know who you are but certainly you are going to a famous blogger if you aren’t
already
Review my website … Bedrock Restoration of Edina
Hello There. I found your weblog the usage of msn. This
is an extremely neatly written article. I’ll be sure to bookmark it and return to read more of your helpful information.
Thanks for the post. I’ll definitely return.
Look into my page; SURFACE PRO REFINISHING
Greetings from Colorado! I’m bored at work so I decided to browse your site on my iphone during lunch break.
I enjoy the info you provide here and can’t wait to take a look when I get home.
I’m shocked at how quick your blog loaded on my phone ..
I’m not even using WIFI, just 3G .. Anyhow, awesome blog!
My web blog: webpage
of course like your web site however you have to test the spelling on quite a
few of your posts. A number of them are rife with spelling issues and
I in finding it very troublesome to inform the reality nevertheless I will certainly come back
again.
my homepage: Bathtub resurfacing 30303
I am curious to find out what blog system you are utilizing?
I’m having some minor security problems with my latest site and I would like to find something more safeguarded.
Do you have any recommendations?
Also visit my blog … Houston ac repair
Thanks for some other excellent article. The place else could anyone
get that kind of info in such a perfect means of writing?
I have a presentation subsequent week, and I am on the search for such info.
Feel free to surf to my homepage; ac repair company
Wow that was odd. I just wrote an really long comment
but after I clicked submit my comment didn’t appear.
Grrrr… well I’m not writing all that over again. Anyhow, just
wanted to say fantastic blog!
Feel free to surf to my blog post :: roofing companies near me
Keep this going please, great job!
Look at my web blog … ac replacement nearby
Write more, thats all I have to say. Literally, it seems as though you relied on the video to
make your point. You clearly know what youre talking about, why waste your intelligence on just posting
videos to your weblog when you could be giving us something enlightening to
read?
Here is my webpage :: Fort Worth Roofers
Hi my family member! I want to say that this post is awesome, nice
written and include almost all vital infos. I would like to peer extra posts like this .
Also visit my homepage :: Fort Worth roof inspection
В обзорной статье вы найдете собрание важных фактов и аналитики по самым разнообразным темам. Мы рассматриваем как современные исследования, так и исторические контексты, чтобы вы могли получить полное представление о предмете. Погрузитесь в мир знаний и сделайте шаг к пониманию!
Подробнее – https://vyvod-iz-zapoya-1.ru/
Do you have a spam issue on this blog; I also am a blogger, and
I was wanting to know your situation; many of us have created some
nice procedures and we are looking to trade strategies with
other folks, be sure to shoot me an e-mail if interested.
My web-site: READY ROOF Inc.
Hmm is anyone else having problems with the pictures on this blog loading?
I’m trying to figure out if its a problem on my end or if it’s the blog.
Any responses would be greatly appreciated.
Also visit my web-site … The Deck Company Custom Deck Builder Nixa MO
Today, I went to the beach front with my children. I found a sea
shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put
the shell to her ear and screamed. There was a hermit crab inside
and it pinched her ear. She never wants to go back! LoL I know this
is completely off topic but I had to tell someone!
My web page; Surface Pro Bathtub repair near me
Amazing blog! Is your theme custom made or did you download it from somewhere?
A theme like yours with a few simple adjustements would really make my blog shine.
Please let me know where you got your theme. Appreciate it
Here is my website :: Marion air conditioning companies
Very good post. I definitely appreciate this site.
Continue the good work!
Feel free to visit my webpage … web page
Your way of explaining the whole thing in this piece of writing is actually good,
every one can without difficulty know it, Thanks
a lot.
my site; web page
Oh my goodness! Impressive article dude! Thanks, However I am having issues
with your RSS. I don’t know the reason why I cannot join it.
Is there anybody having similar RSS problems? Anyone who knows the answer will
you kindly respond? Thanks!!
Take a look at my blog shower refinishing near me
Great beat ! I wish to apprentice while you amend your website,
how could i subscribe for a weblog website? The account aided me a applicable deal.
I have been a little bit familiar of this your broadcast provided bright
transparent idea
Feel free to surf tto my blog post :: Buy Best SEO Backlinks
I just couldn’t depart your site before suggesting that I extremely enjoyed the standard information a person provide on your visitors?
Is gonna be again regularly to investigate cross-check new posts
Also visit my homepage Suburban local sump pump installation near me
Have you ever considered writing an e-book or guest
authoring on other websites? I have a blog based upon on the same ideas you discuss
and would love to have you share some stories/information. I know my audience would appreciate your work.
If you’re even remotely interested, feel free to shoot me an e-mail.
Visit my blog post :: ac company
I do not even know how I ended up here, but I thought this post was great. Cheers!
Cheers!
I do not know who you are but certainly you’re going to a famous blogger
if you are not already
my web site: webpage
Hmm it seems like your blog ate my first comment (it was
extremely long) so I guess I’ll just sum it up what I wrote
and say, I’m thoroughly enjoying your blog.
I too am an aspiring blog writer but I’m still new to the whole
thing. Do you have any tips and hints for first-time blog writers?
I’d certainly appreciate it.
My blog :: homepage
I do not even know the way I ended up right here, however I thought this submit used
to be great. I don’t understand who you might be but certainly you’re going to a
well-known blogger for those who aren’t already.
Cheers!
Here is my website :: Foster Plumbing & Heating
пройти психиатра платного психиатра [url=http://psihiatry-nn-1.ru]пройти психиатра платного психиатра[/url] .
Very good article. I definitely appreciate this
site. Thanks!
Feel free to surf to my page – HVAC Services Near Me
Great post. I was checking continuously this blog and I’m impressed! I care for
I care for
Extremely useful info specially the last part
such info much. I was seeking this certain info for a long time.
Thank you and good luck.
my web page – Local Commercial Power Washing Company
I’m extremely impressed with your writing skills as well as
with the layout on your blog. Is this a paid theme or
did you modify it yourself? Anyway keep up the nice quality writing, it’s rare to see a great blog
like this one today.
Feel free to surf to my website … BDE Construction & Kitchen Cabinets
Wow, fantastic blog layout! How lengthy have you ever been running a blog for?
you made running a blog look easy. The overall look of your site is excellent, as smartly as the content material!
my web page; web page
With havin so much content and articles do you ever
run into any problems of plagorism or copyright violation? My site has a lot of unique content I’ve either created
myself or outsourced but it looks like a lot of it is
popping it up all over the internet without my permission. Do
you know any techniques to help reduce content from being ripped off?
I’d certainly appreciate it.
my web site :: neck pain relief Boise
Wonderful beat ! I wish to apprentice while you amend
your web site, how can i subscribe for a blog site? The account helped me
a acceptable deal. I had been a little bit acquainted of this your broadcast
offered bright clear idea
Stop by my page :: vet services
I quite like reading an article that can make people think.
Also, many thanks for allowing for me to comment!
Also visit my blog … water softener installation
My spouse and I stumbled over here coming from a different web address and
thought I might as well check things out. I like what I see so i am just following you.
Look forward to exploring your web page repeatedly.
Here is my web site: drain cleaning services
Just desire to say your article is as surprising.
The clearness in your post is just cool and i can assume you are an expert on this subject.
Well with your permission allow me to grab your feed to keep
up to date with forthcoming post. Thanks a million and please keep up the gratifying work.
Feel free to visit my web-site … Ready best roof replacement services
Hello there! This post couldn’t be written any better!
Reading this post reminds me of my old room mate! He always kept
talking about this. I will forward this article to him.
Pretty sure he will have a good read. Thanks for sharing!
My site :: Personal Injury Lawyer near me
I was wondering if you ever thought of changing the layout of
your site? Its very well written; I love what youve got
to say. But maybe you could a little more in the way of content so people
could connect with it better. Youve got an awful lot
of text for only having 1 or two pictures. Maybe you could space it out
better?
Check out my web-site – veterinary clinic
I don’t even understand how I finished up right here, but I assumed this post used to be great.
I don’t know who you are however definitely you are going
to a famous blogger when you are not already.
Cheers!
Also visit my web site :: splash aqua park
Pretty! This was an extremely wonderful article. Thanks for providing these details.
Review my website; water softener installation
Ahaa, its nice discussion about this piece of writing here at this web site,
I have read all that, so at this time me also commenting at this place.
my blog post; auto tint near me
This is a topic which is close to my heart… Thank you!
Where are your contact details though?
Here is my web page … attorney
My brother recommended I might like this website.
He was totally right. This post truly made my day.
You can not imagine simply how much time I had spent for this information! Thanks!
My blog; corporate event venues near me
Thanks for another fantastic article. The place else could anybody get that kind of info in such an ideal approach of writing?
I have a presentation subsequent week, and I’m at the look for such information.
Here is my web site Somers Plumbers – Phoenix Plumbing Company
Hurrah, that’s what I was exploring for, what
a stuff! present here at this blog, thanks admin of this web site.
my blog Poker & DominoQQ Online
Hello, Neat post. There is a problem together with your web site in web explorer, could check this?
IE nonetheless is the marketplace chief and a large component of other people will omit your great writing because of this problem.
My blog post :: wedding venues long island
If you desire to grow your knowledge simply keep visiting this
website and be updated with the most up-to-date news update posted here.
Here is my webpage; painting service near me
Please let me know if you’re looking for a article writer for
your blog. You have some really good articles and I think I would be a good asset.
If you ever want to take some of the load off, I’d absolutely love
to write some material for your blog in exchange for a link back to mine.
Please send me an e-mail if interested. Kudos!
Also visit my page: bathroom remodeling company
I don’t even know how I ended up here, but I thought this post was Cheers!
Cheers!
great. I do not know who you are but certainly you are going to a famous blogger if you are not already
Here is my web blog :: stair railing painting
Unquestionably believe that which you said. Your favorite justification appeared to be on the web the easiest factor to take into account of.
I say to you, I certainly get irked even as other
people consider issues that they just don’t realize about.
You controlled to hit the nail upon the top and outlined out the whole thing with no need side-effects , people
could take a signal. Will likely be again to get more.
Thanks
Feel free to visit my website :: Best window tinting IA
This is the right webpage for anybody who hopes to understand this
topic. You understand so much its almost hard to argue with
you (not that I actually would want to…HaHa). You certainly
put a fresh spin on a topic that has been written about for
years. Wonderful stuff, just excellent!
Also visit my website; water damage restoration services near me
An impressive share! I’ve just forwarded this
onto a friend who has been conducting a little homework on this.
And he actually ordered me breakfast because I found it
for him… lol. So let me reword this….
Thank YOU for the meal!! But yeah, thanx for spending the
time to talk about this issue here on your web page.
Also visit my page; Lawn care West Des Moines
Thanks in support of sharing such a nice idea, piece of writing is fastidious, thats why i have read
it entirely
Feel free to visit my web page: custom deck builder near me
I think what you published made a great deal of sense.
But, what about this? suppose you were to
create a killer headline? I am not saying your information isn’t good, however suppose you added a title that makes people desire more?
I mean Rosetta Cartesian_ddG: 更快更准确的蛋白质稳定性预测方法 – BioEngX is kinda vanilla.
You could glance at Yahoo’s front page and watch how
they create news headlines to grab people interested. You might try adding a video or a picture or two to grab people interested about what you’ve written. In my opinion, it might bring your website a little
livelier.
Feel free to visit my web blog :: Paver seal coating
Its like you read my thoughts! You seem to understand
a lot about this, like you wrote the ebook in it or something.
I feel that you simply can do with some percent
to pressure the message home a little bit, however other than that, this is excellent blog.
An excellent read. I will certainly be back.
My blog post: Facebook Proxies
I think this is one of the most significant info for me.
And i am glad reading your article. But want to remark on some general things,
The website style is great, the articles is really great : D.
Good job, cheers
Feel free to visit my homepage :: local cabinet maker
Excellent post. I used to be checking continuously I take care of
I take care of
this weblog and I’m impressed! Extremely useful info specifically the last section
such info a lot. I was seeking this particular information for a very long time.
Thank you and good luck.
my site; homepage
What’s up to every one, the contents present at this web page are
actually remarkable for people knowledge, well, keep up the nice work fellows.
Also visit my webpage gutter replacement
Valuable info. Lucky me I found your site by chance, and I
am shocked why this coincidence did not came about in advance!
I bookmarked it.
Feel free to visit my page; animal hospital Greensburg
I’ll immediately take hold of your rss as I can’t find your
email subscription link or newsletter service. Do you’ve any?
Kindly allow me recognize so that I may just subscribe.
Thanks.
Feel free to surf to my web site Sewage Cleanup near me
I am regular visitor, how are you everybody? This piece of writing posted at this web site is
actually good.
Here is my web-site … Water Heater Repair
I’m not sure why but this weblog is loading incredibly slow for me.
Is anyone else having this problem or is it a issue on my end?
I’ll check back later and see if the problem still exists.
Feel free to surf to my page: concrete contractors
I like the valuable info you provide on your articles. I’ll bookmark
your weblog and test again right here frequently.
I am moderately certain I will be told plenty of new stuff
right right here! Good luck for the next!
my website :: dumpster contractor
Very nice post. I just stumbled upon your blog and wished to
say that I have really enjoyed browsing your blog posts.
In any case I will be subscribing to your rss feed and I hope you write again soon!
My page Dedicated Private Proxy
Way cool! Some very valid points! I appreciate you penning this post and also the rest of the website is very good.
Also visit my web blog; Peak Performance Restoration
First off I would like to say superb blog! I had a quick question in which I’d like to ask if you do not mind.
I was interested to know how you center yourself and clear your
thoughts prior to writing. I have had trouble clearing my thoughts in getting my thoughts out.
I do enjoy writing however it just seems like the first 10 to
15 minutes tend to be wasted simply just trying to figure out how to begin. Any suggestions or
hints? Kudos!
Also visit my web blog … Burks and Company
Hi there! Quick question that’s completely off topic.
Do you know how to make your site mobile friendly? My blog looks weird when viewing from my iphone.
I’m trying to find a theme or plugin that might be able to correct this problem.
If you have any recommendations, please share. Many thanks!
Here is my web-site – Situs Bola Tangkas
Hello, everything is going nicely here and ofcourse every one is sharing facts,
that’s truly good, keep up writing.
Here is my blog post: Around the Town Heating and Cooling
It’s really a great and useful piece of information. I
am glad that you just shared this useful info with
us. Please stay us up to date like this. Thanks for sharing.
my blog Burks and Company
Hi I am so grateful I found your blog page, I really found you by accident, while I was
researching on Askjeeve for something else, Anyhow I am here now and
would just like to say kudos for a fantastic post and a all
round thrilling blog (I also love the theme/design), I don’t have time to
read through it all at the minute but I have book-marked
it and also included your RSS feeds, so when I have
time I will be back to read a lot more, Please do keep up
the fantastic jo.
my webpage: LeTourneau & Family Orthotics and Prosthetics
Hi Dear, are you genuinely visiting this site on a regular basis,
if so after that you will definitely get pleasant knowledge.
Feel free to visit my webpage … mold remediation services
This paragraph provides clear idea in favor of the new users of blogging, that truly how to do blogging.
Feel free to visit my web page: voyance immédiate sans cb
I read this article fully regarding the comparison of newest and previous technologies, it’s amazing article.
Here is my web-site :: homepage
If you want to obtain a good deal from this paragraph then you have to apply such strategies to
your won website.
My web blog; Force 1 Restoration Services
Every weekend i used to pay a quick visit this web page, for the reason that i want enjoyment, since
this this web site conations truly fastidious funny material too.
my blog post – Quality Heating and Air
ini adalah situs bokep paling bagus
My web-site situs porno
Hey! Quick question that’s totally off topic. Do you
know how to make your site mobile friendly? My weblog looks
weird when viewing from my iphone 4. I’m trying to find a theme or plugin that might be able to correct this problem.
If you have any recommendations, please share.
Thanks!
Here is my web-site … dominoqq
Hey I know this is off topic but I was wondering if you knew of any widgets
I could add to my blog that automatically tweet my newest twitter updates.
I’ve been looking for a plug-in like this for quite some time and
was hoping maybe you would have some experience
with something like this. Please let me know if you run into
anything. I truly enjoy reading your blog and I look forward to your new updates.
Also visit my blog :: https://telegramisrael.com/
Hello there! I know this is kinda off topic but I was wondering which blog platform
are you using for this site? I’m getting sick and tired of WordPress because I’ve had problems with hackers and I’m
looking at alternatives for another platform. I would be great if you could point
me in the direction of a good platform.
my web site … LeTourneau & Family Orthotics and Prosthetics
I am sure this piece of writing has touched all the
internet viewers, its really really good post on building up new website.
Feel free to visit my web page … An Honest Day’s Power Washing
Hi, its pleasant paragraph on the topic of media print, we all be familiar
with media is a wonderful source of information.
Check out my web site :: Force 1 Restoration Services
Hello, I read your blogs regularly. Your story-telling style is awesome,
keep it up!
Take a look at my page Social Networks Backlinks
I do not even know the way I ended up here, but I assumed this publish was good.
I don’t know who you’re however certainly you are going
to a well-known blogger for those who are not already.
Cheers!
Also visit my page … Southern Iowa Plumbing
Right away I am going away to do my breakfast, afterward having my breakfast
coming again to read more news.
my site … Around the Town Heating and Cooling
Very nice write-up. I definitely appreciate this site.
Thanks!
Here is my page; Bandar Taruhan Bola & Casino Online dengan Pasaran Lengkap
Hi there! I’m at work browsing your blog from my new iphone!
Just wanted to say I love reading your blog and look forward to all your posts!
Keep up the outstanding work!
Feel free to surf to my web-site … Around the Town Heating and Cooling
Aw, this was a very good post. Taking a few minutes and actual effort
to make a really good article… but what can I say… I hesitate a whole lot and don’t manage to get nearly anything done.
My web site: Mighty Movers Logistics
I am sure this article has touched all the internet visitors, its really really good paragraph on building
up new web site.
My web page :: Affordable Tree & Lawn Service LLC
I love your blog.. very nice colors & theme. Did you create this website yourself or did you
hire someone to do it for you? Plz answer back as I’m looking to create my own blog and would like
to find out where u got this from. thanks
Here is my site Quality Heating and Air
I got this web site from my friend who told me about this website and at the moment this time I am browsing this website
and reading very informative articles here.
Look at my web site … Around the Town Heating and Cooling
Thanks for your personal marvelous posting! I seriously enjoyed reading it, you may be a great author.
I will make certain to bookmark your blog and will come
back from now on. I want to encourage you continue your great writing,
have a nice holiday weekend!
Here is my page: Mighty Movers Logistics
This is my first time pay a visit at here and i am actually
pleassant to read all at alone place.
Here is my blog post – hydro jet drain cleaning
Have you ever considered about including a little bit more than just your articles?
I mean, what you say is valuable and all. However just imagine if you added some great pictures or video clips to give your
posts more, “pop”! Your content is excellent but with
images and video clips, this website could definitely be one
of the very best in its niche. Very good blog!
Also visit my webpage: Quality Heating and Air
I’m not sure exactly why but this site is loading
very slow for me. Is anyone else having this problem or is it a problem on my end?
I’ll check back later and see if Around the Town Heating and Cooling problem still exists.
Superb, what a website it is! This webpage gives helpful data to us,
keep it up.
Feel free to surf to my website: LeTourneau & Family Orthotics and Prosthetics
Woah! I’m really loving the template/theme of this site.
It’s simple, yet effective. A lot of times it’s hard to get that “perfect balance” between usability and visual appearance.
I must say you’ve done a excellent job with this. Additionally, the blog loads super
fast for me on Safari. Superb Blog!
Here is my web-site; water heater replacement near me
Hello! Do you know if they make any plugins to safeguard against hackers?
I’m kinda paranoid about losing everything I’ve worked hard on. Any suggestions?
my homepage Southern Iowa Plumbing
Hi there, after reading this amazing piece of writing i am also delighted
to share my know-how here with friends.
Feel free to visit my page voyance immédiate (Trent)
This is my first time pay a visit at here and i am really pleassant to read everthing at
alone place.
My blog post DRYmedic Restoration Services of Baton Rouge
I think the admin of this site is actually working hard in favor of his web page, since here every information is quality based information.
Also visit my website … consultation voyance téléphone (Ernestina)
situs porno yang paling bagus
Feel free to surf to my website; situs video porno
Hey! I just wanted to ask if you ever have any problems with hackers?
My last blog (wordpress) was hacked and I ended up losing months of hard work due to no data backup.
Do you have any methods to prevent hackers?
Here is my web blog Plumbing Services of Raleigh
I have read so many articles or reviews regarding the blogger
lovers however this piece of writing is actually a fastidious piece of writing, keep it
up.
My site: An Honest Day’s Power Washing
I blog frequently and I genuinely thank you for your content.
This article has really peaked my interest. I’m going to bookmark your website and keep checking for new details about once per week.
I subscribed to your Feed as well.
Feel free to visit my website … An Honest Day’s Power Washing
Sweet blog! I found it while surfing around on Yahoo News.
Do you have any suggestions on how to get listed
in Yahoo News? I’ve been trying for a while but
I never seem to get there! Thanks
Feel free to surf to my web blog :: טלגראס כיוונים במזכרת בתיה – https://telegrass2u.com/,
I just could not depart your web site prior to suggesting that I
extremely loved the usual information an individual supply in your guests?
Is gonna be back incessantly to investigate cross-check new posts
Feel free to surf to my web site Plumbing Services of Raleigh
What’s up Dear, are you truly visiting this website regularly, if
so after that you will absolutely take fastidious know-how.
Have a look at my web blog Plumbing Services of Raleigh
Hello to all, how is the whole thing, I think every one is getting more
from this web page, and your views are good in support of
new visitors.
Feel free to surf to my web-site … An Honest Day’s Power Washing
ini adalah situs bokep paling bagus
Also visit my web page bokep indonesia
Hey!
I’m Tom, and I really wanted to leave a comment here.
Your blog is inspiring, and it’s always nice to
come across such cool posts.
By the way, if you’re curious about fun chats, you should definitely
check out Bubichat! It’s an amazing text chat
where fun conversations happen all the time.
I spend so much time on it because it’s so easy to meet someone special and enjoy private chats.
Give it a try if you’re up for something different!
I’m usually there, and who knows – we might chat soon!
Can’t wait to see more from you! adult chat with strangers
XOXO Waiting on bubichat.com
I really like your blog.. very nice colors & theme.
Did you create this website yourself or did
you hire someone to do it for you? Plz reply as I’m looking to construct
my own blog and would like to find out where u got this from.
thank you
Feel free to surf to my web page … roofing company houston services
situs bom88 situs link bokep yang banyak tersedia berbagai macam jenis video bokep yang ada
Also visit my web page :: bom88 situs anjing
Very good information. Lucky me I ran across your website by chance (stumbleupon).
I’ve book-marked it for later!
my homepage … Water damage restoration contractor near me
Wonderful beat ! I would like to apprentice even as you amend your
site, how could i subscribe for a weblog website?
The account helped me a applicable deal. I had been tiny bit
familiar of this your broadcast offered shiny clear idea
Also visit my website – Best private proxies
situs bokep bom88 situs bokep aw aw ase ase
asek kali
Fine way of telling, and nice piece of writing to
take data concerning my presentation subject matter, which i am
going to present in school.
Here is my blog post; roof cleaning near me
Great post. I was checking constantly this blog and I’m I care for such info a lot. I was seeking this particular information for a very long time.
I care for such info a lot. I was seeking this particular information for a very long time.
impressed! Extremely useful info specifically the
last part
Thank you and good luck.
Also visit my blog post; Residential roofing
Hi friends, fastidious piece of writing and pleasant urging
commented at this place, I am genuinely enjoying by these.
Stop by my homepage 100 Proxies
of course like your web-site but you need to take a
look at the spelling on several of your posts. A number of them are
rife with spelling issues and I in finding it very bothersome to inform the
reality however I will surely come back again.
Stop by my web blog; Situs asli dan resmi Tangkasnet
It’s in fact very difficult in this busy life to listen news on TV,
therefore I only use world wide web for that purpose, and get the newest information.
Feel free to surf to my page – PuroClean of Rahway
Terrific work! That is the type of info that should be shared around the net.
Shame on Google for no longer positioning this publish upper!
Come on over and discuss with my site . Thanks =)
my site: Best DA 50 Backlinks
I always emailed this blog post page to all my contacts, because if like to read it then my friends
will too.
Here is my web page; agen Bola Tangkas
I could not refrain from commenting. Well written!
Here is my page :: Buy Proxies
Hello my family member! I want to say that
this post is awesome, great written and come with almost all important infos.
I’d like to peer more posts like this .
my web page: Flood Damage Restoration service
Ahaa, its pleasant discussion concerning this paragraph at this
place at this website, I have read all that, so now me also commenting here.
Take a look at my web blog: Proxy price
Thanks for your marvelous posting! I quite enjoyed reading it, you can be
a great author.I will make sure to bookmark your blog and definitely will come back someday.
I want to encourage yourself to continue your great writing,
have a nice weekend!
Here is my blog :: web page
Pretty nice post. I just stumbled upon your blog and wished to say that
I have really enjoyed surfing around your blog posts.
After all I’ll be subscribing to your rss feed and
I hope you write again very soon!
Stop by my blog post; home interior remodeling
Greetings from California! I’m bored to tears at work so I decided to browse your site on my iphone during lunch
break. I really like the information you present here and can’t wait to take a look when I get
home. I’m shocked at how fast your blog loaded on my
mobile .. I’m not even using WIFI, just 3G .. Anyhow, wonderful site!
Feel free to surf to my web-site web site
Hi there, just became alert to your blog through Google, and found that it’s really informative.
I’m going to watch out for brussels. I will appreciate if you continue this in future.
Numerous people will be benefited from your writing.
Cheers!
Feel free to visit my blog; Instagram Proxies
I truly love your website.. Pleasant colors & theme.
Did you develop this site yourself? Please reply back as I’m
planning to create my own site and would love to find out where you
got this from or what the theme is called.
Appreciate it!
Have a look at my web blog – roof replacement
Thanks for the good writeup. It in fact was a enjoyment
account it. Look complex to far brought agreeable from you!
By the way, how could we keep in touch?
Also visit my webpage :: vibely diamond mascara review
WOW just what I was looking for. Came here by searching for
A Perfect Finish Painting
Here is my web site commercial painting service
I’m really enjoying the design and layout of your blog.
It’s a very easy on the eyes which makes it much more enjoyable for me to come here and visit more often. Did
you hire out a designer to create your theme? Excellent work!
My web-site: jasa top up saldo Namecheap
I was able to find good advice from your content.
Here is my blog post … Painting
Hurrah! After all I got a blog from where I can really take helpful data concerning my study and
knowledge.
Here is my webpage; Get Free Proxy Ip Address
My family always say that I am killing my time here at net, but I know
I am getting know-how daily by reading thes fastidious content.
Also visit my web blog is vibely mascara cruelty free
Ayo mainkan peluang besar anda di LIGACOR untuk kaya dengan bermain game
di website terbaik tanpa stress! Dapatkan info cuan mudah dan jadilah kaya secara otomatis
I am regular visitor, how are you everybody? This paragraph posted at this website is in fact
good.
Stop by my web site Woodbury Carpet Stretching
Thank you for every other informative web site. Where else may I get that kind of information written in such an ideal manner?
I have a project that I’m simply now running on, and
I have been at the glance out for such information.
Also visit my web blog – penyedia jasa Bitcoin
Aw, this was an extremely nice post. Finding the
time and actual effort to create a top notch article… but what can I say… I
procrastinate a whole lot and never seem to get nearly anything
done.
Here is my homepage :: Mold Remediation nearby
Apresiasi, Beragam konten yang sangat bermanfaat!
My web-site; Layanan Pendirian Perseroan Terbatas Terpercaya bersama RuangOffice.
What’s up friends, nice article and fastidious arguments commented here,
I am in fact enjoying by these.
Feel free to surf to my blog – homepage
Picuki is a powerful software for these who value privateness, allowing users to browse Instagram anonymously.
Feel free to visit my homepage: tiktok viewer
This article will assist the internet visitors for creating new
web site or even a blog from start to end.
Check out my homepage: reviews on vibely mascara
This is very interesting, You’re a very skilled blogger.
I’ve joined your rss feed and look forward to seeking more of
your excellent post. Also, I’ve shared your site in my
social networks!
My website pool floor designs
LIGACOR ♦️♦️ LINK LOGIN ALTERNATIF SITUS SLOT AUTO MAXWIN DIJAMIN TERPERCAYA SAAT INI YANG
MENAWARKAN PLATFORM PERMAINAN YANG AMAN, TERPERCAYA, DAN INOVATIF, SERTA BONUS MENARIK DAN LAYANAN PELANGGAN 24/7.
2024
The other day, while I was at work, my cousin stole my iphone
and tested to see if it can survive a 30 foot drop, just so
she can be a youtube sensation. My apple ipad is now
destroyed and she has 83 views. I know this is entirely
off topic but I had to share it with someone!
Also visit my web site – marketing [Raina]
Hello there! This is my first visit to your blog!
We are a collection of volunteers and starting a new project in a community in the same niche.
Your blog provided us valuable information to work on. You have done a
outstanding job!
my web page … vibely xpress control mascara reviews
Wow, this article is nice, my younger sister is analyzing
these things, therefore I am going to tell her.
Feel free to surf to my blog :: vibely mascara australia
Saved as a favorite, I love your website!
Have a look at my web site; where is vibely mascara made
Thank you for another great article. Where else may anybody get that
kind of information in such a perfect approach of writing?
I have a presentation subsequent week, and I’m at the look for such information.
Check out my webpage: vibely vs thrive mascara
Spot on with this write-up, I honestly believe this web site needs far more
attention. I’ll probably be returning to see more,
thanks for the info!
Also visit my web-site – lush cosmetics vibely mascara
Hello colleagues, its wonderful piece of writing regarding
cultureand fully defined, keep it up all the time.
Feel free to surf to my web site :: is vibely mascara waterproof
Having read this I thought it was very enlightening. I appreciate you finding the time and energy to put this information together. I once again find myself personally spending a significant amount of time both reading and posting comments. But so what, it was still worthwhile!
LIGACOR adalah
platform terpercaya untuk login, daftar, dan mengunduh
aplikasi resmi. Dapatkan akses cepat dan aman dengan fitur terbaik hanya di sini
It’s appropriate time to make some plans for the
long run and it is time to be happy. I have learn this submit
and if I may I desire to suggest you few attention-grabbing issues or tips.
Maybe you could write subsequent articles regarding this article.
I wish to read even more issues approximately it!
Visit my website :: vibely mascara reviews
Hello, after reading this amazing article i am too glad to share my familiarity here with mates.
My web-site Buy 10000 Private Proxies
It’s actually a cool and useful piece of information. I am glad that you just shared this helpful info with us.
Please stay us up to date like this. Thanks for sharing.
my web-site … best Quality Seo Backlinks
(seobests.Com)
This is very interesting, You’re a very skilled blogger.
I’ve joined your rss feed and look forward to seeking more of your magnificent
post. Also, I have shared your website in my social networks!
Feel free to visit my site Private proxies [privateproxies.top]
This is really interesting, You are a very skilled blogger.
I have joined your rss feed and look forward to seeking more of your great post.
Also, I have shared your website in my social networks!
Also visit my web blog – Buy 200 Cheap Private Proxies
I don’t even understand how I stopped up here, however
I assumed this publish was good. I do not recognise who you are however definitely you
are going to a famous blogger if you happen to aren’t already.
Cheers!
my web site – Best SEO Backlinks
Thanks for the marvelous posting! I really enjoyed reading it,
you can be a great author. I will be sure to bookmark your blog
and will come back from now on. I want to encourage you to continue your great work, have
a nice evening!
Also visit my web site Cheap Proxies
Can I simply just say what a relief to discover someone that
truly understands what they’re talking about on the
net. You definitely realize how to bring a problem to light and make
it important. A lot more people should check
this out and understand this side of the story. I can’t believe you aren’t more popular since you certainly possess the gift.
My webpage: 400 Private Proxies
Hello there! This is my first visit to your blog! We are a team
of volunteers and starting a new project in a community in the same
niche. Your blog provided us beneficial information to
work on. You have done a outstanding job!
My web blog; 1000 Cheap Proxies
excellent points altogether, you simply won a new reader. What might you recommend in regards to your put up that you simply made a few days in the past?
Any certain?
Feel free to visit my blog Buy 100 Cheap Private Proxies
I’m not that much of a internet reader to be honest
but your sites really nice, keep it up! I’ll
go ahead and bookmark your website to come back down the road.
Cheers
my page … Dedicated Proxy
Elizabeth Holmes, quien fuera la CEO de Theranos, enfrentó una sentencia que capturó la atención del mundo tecnológico.
my site noticias sobre tecnologia
Hello there, You have done an incredible job.
I will certainly digg it and personally recommend to my friends.
I’m confident they’ll be benefited from this website.
Also visit my site – private proxies usa (maxiproxies.com)
I think the admin of this site is actually working hard in support of his web page, for the reason that here every material is quality based data.
cialis us – order viagra 100mg without prescription cheap sildenafil for sale
sildenafil pills 25mg – order cialis 10mg brand tadalafil
Następnie wróć do getmyfb.com i wklej link w polu tekstowym na stronie głównej.
Feel free to surf to my webpage; facebook downloader
You actually stated this really well!
golden nugget online casino phone number https://linkscasino.info/tennis-betting/ besplatne casino igre online
バイアグラ通販 安全 – г‚·г‚ўгѓЄг‚№ гЃЉгЃ™гЃ™г‚Ѓ г‚їгѓЂгѓ©гѓ•г‚Јгѓ«гЃ®йЈІгЃїж–№гЃЁеЉ№жћњ
adana halı yıkama karbeyaz halı yıkama adana
Asking questions are really fastidious thing if you are not understanding
something completely, except this piece of writing provides nice understanding even.
гѓ—гѓ¬гѓ‰гѓ‹гѓігЃ®йЈІгЃїж–№гЃЁеЉ№жћњ – гѓ—гѓ¬гѓ‰гѓ‹гѓігЃ®иіје…Ґ г‚ёг‚№гѓгѓћгѓѓг‚ЇйЂљиІ©гЃЉгЃ™гЃ™г‚Ѓ
¡SSSPornHub es tu mejor opción para contenido de
video de alta calidad!
Feel free to visit my webpage; pornhub downloader
Cheers! Lots of material.
online casino schweiz legal las vegas online casino real money how many casino missions are there in gta online
cozaar 50mg pills – cephalexin without prescription cost cephalexin 500mg
Nicely put, Regards.
online casino geld zurГјck sofortГјberweisung casino slots online free casino nj online
With thanks! Loads of facts.
bonifico bancario casino online casino canada real casino online usa
You said it adequately.!
nytt casino online online real money casino vegas lounge online casino
You said it adequately..
casino gran madrid poker online casino slots online real money agent online casino malaysia
Amazing write ups. Thanks.
888 online casino michigan real money casino canada online online usa casinos real money
Whoa a lot of great facts.
skycity online casino paysafecard casino games online for real money nuevos casinos online en mexico
You actually revealed this exceptionally well!
best bank transfer online casino in india casino online usa real money top online casinos germany
This is nicely put! !
new ontario online casinos best online casino sites canada big bass bonanza online casino
Nicely put. Appreciate it!
play live casino games online new online casino no deposit bonus usa online online casino
Amazing quite a lot of excellent facts.
demo casino online online casino slots best match bonus online casino
This is nicely put! !
versailles gold casino game online best online casino canada real money 333 casino online
buy augmentin for sale – brand clavulanate levoxyl oral
This is nicely said. .
online mobile casino no deposit bonus online casino list america’s best online casinos
This is nicely said. .
online casinos like chumba best online real money casino legit online casino free spins
Information well taken.!
stardust online casino promo code best casinos online canada what’s the best online casino to win money
Wow all kinds of amazing info!
777 casino play online real money online casinos party casino online
Thanks a lot, I appreciate it.
online casino deposit $1 online casino top rated online casinos zonder vergunning
Amazing many of great knowledge!
casino online lớn nhất thế giới online casino with free signup bonus real money usa legal online casinos in south africa
Amazing advice. Many thanks.
best australian casino online casino online real money usa bally online casino blackjack
Regards. A lot of posts.
parx casino online pa real money casino online 747 live online casino philippines
Excellent facts. Appreciate it!
bingo hall online casino login new usa online casinos online casino mobil
Thanks! I like it!
alberta online casino casino slots online real money ada online casinos
Really quite a lot of superb tips!
bonanza online casino casino real money online ver casino royale online gratis
This design is steller! You obviously know how to keep a reader amused.
Between your wit and your videos, I was almost moved
to start my own blog (well, almost…HaHa!) Great job.
I really enjoyed what you had to say, and more than that, how you presented it.
Too cool!
My site JUDI ONLINE
Nicely put, Thanks a lot!
neue online casinos 2024 online casino games that pay real money andromeda online casino
“Was it because of that?” His dad asks as he points a finger towards him. And his midsection.
Great stuff. Thank you.
mgm online casino down casino online usa real money best online casino apps
Whoa quite a lot of fantastic info.
best online casino in finland party casino canada new usa online casinos 2022 real money
Very good tips. Thank you.
winstar online social casino best payout online casino real money igt online casino real money
Whoa all kinds of excellent data.
energy online casino online casino play for real money malaysia online casino free credit for new member
Regards! I appreciate it.
are online casinos regulated best online casino slot machines 10 mejores casinos online
I am curious to find out what blog platform you are working
with? I’m experiencing some minor security issues with my latest blog
and I’d like to find something more safe. Do you have any recommendations?
Review my website :: slot gacor bonus deposit
buy betnovate cheap – order benoquin for sale buy monobenzone without a prescription
This is really interesting, You’re a very skilled blogger.
I have joined your feed and look forward to seeking more of your excellent post.
Also, I have shared your web site in my social networks!
Here is my site JUDI ONLINE
“I look forward to the day when you are a fully grown man, my son.” His father says. “I am anxious to see the fruit of my loins reach his fully-adorned manhood. Adulthood.” “That’s it! That’s it! That’s it!” His father bellows.
deltasone 5mg oral – buy generic prednisone online order elimite for sale
purchase absorica online cheap – dapsone usa deltasone for sale online
“Once you plant that cock of yours in some squirmy hole and empty those warm balls of yours into a moist wet hole. Then you are on your way to becoming a man, and only then will you, come-of-age. But it is only a step onto the winding pathway towards manhood. It is my job to teach you what it means to be a man.” His father stands as he finishes his sentence. Loosening his belt and pulling his shirt out from the tucked confines of his pants. “I ain’t a-talking ‘bout how big ya cock is, boy. I am talking ‘bout ya, weight. Your height.” His dad says. “I kinda figured on how big you are down there. That’s obvious. It makes me proud of you. My son. Of what I created, along with ya mother.”
Thank you for sharing your info. I really appreciate your efforts and I will be waiting for your further write
ups thanks once again.
Look at my web blog … JUDI ONLINE
Hi there, just became aware of your blog through Google,
and found that it’s really informative. I am gonna watch out for brussels.
I will be grateful if you continue this in future.
Lots of people will be benefited from your writing.
Cheers!
Take a look at my web site … slot gacor china
Well voiced really. !
casino aladdin online [url=https://usagamblinghub.com/#]us online casino[/url] billionaire casino online
Very good write ups, Cheers!
free online casino slots with bonus rounds [url=https://usacasinomaster.com/#]free casino online[/url] bandar taruhan casino joker123 online
His father’s words are what he hears when he erupts. His cum streaming like liquid threads from the pee-hole of his rigid cock. gay sex porn “Yes, sir.” He says as he straightens up, standing erect as his member pulses to life between his legs, and in his father’s right hand, which are firmly locked on his balls. “Yes, sir, I do.”
Wonderful write ups. Thank you!
aztec riches casino online [url=https://luckyusaplay.com/#]best online usa casino[/url] play online casino free win real money
Truly a good deal of great tips.
william hill casino online [url=https://luckyusaplay.com/#]casino bonus online[/url] encore online casino
I am really inspired with your writing talents as well as with the structure
for your weblog. Is this a paid subject matter or
did you customize it yourself? Either way stay up the nice quality writing, it is
uncommon to peer a nice weblog like this one nowadays..
Also visit my blog – JUDI ONLINE
Hey just wanted to give you a quick heads up. The
words in your article seem to be running off the screen in Chrome.
I’m not sure if this is a format issue or something to do with browser compatibility but I figured I’d
post to let you know. The design look great though!
Hope you get the problem fixed soon. Thanks
Stop by my web-site :: JUDI ONLINE
Hello friends, its fantastic post concerning teachingand entirely explained, keep it up all the time.
Feel free to visit my web site; JUDI ONLINE
Reliable knowledge. Kudos.
chumba casino online mobile casino play online online casino legaal
Thanks, Numerous data.
online casino real money las atlantis best usa online casinos best social casinos online
You stated that perfectly!
mo online casino site live online casino niagara casino online login
You have made your point!
online casino app philippines [url=https://uscasinoguides.com/#]online casino gaming[/url] how to report online casino
Terrific content, Appreciate it!
best 20 deposit casinos in usa to play online play casino online free online casinos usa real money
Thank you! A lot of posts.
top dragonfish online casinos [url=https://usagamblinghub.com/#]free casino online[/url] 1 win casino online
Seriously many of helpful tips!
how to choose online casino online casino with no deposit bonus sa online casinos zar
Great info, Kudos!
list of the best online casinos [url=https://usacasinomaster.com/#]best online casinos that payout[/url] como ganar en casino online
Regards! Useful information!
real casino games online free us online casinos legitimate online casino games
Nicely spoken without a doubt! !
online casino schleswig holstein [url=https://luckyusaplay.com/#]play casino online[/url] peermont online casino
Thanks a lot, I value this!
online casino barriГЁre spiele in der schweiz online casino deposit bonus low minimum deposit casinos online
Thanks a lot! Ample data.
win big online casino [url=https://uscasinoguides.com/#]live dealer online casino[/url] online kostenlos casino
Cheers, Loads of facts!
best new online casino uk online casino no deposit bonus codes online casino international
Amazing write ups. Thanks.
poker star casino online [url=https://usaplayerscasino.com/#]casino gambling online[/url] casino bet online real money
purchase trihexyphenidyl generic – order trihexyphenidyl for sale purchase emulgel for sale
Superb knowledge. Appreciate it!
online casino legit [url=https://usagamblinghub.com/#]play online casino[/url] el royale casino online reviews
Many thanks! A lot of posts!
best usa online casinos with no deposit bonus [url=https://usacasinomaster.com/#]casino online no deposit bonus[/url] horseshoe casino application online
Really plenty of useful material!
hollywood online casino best online casino usa caesars casino pa online
Regards. I appreciate this.
online casino paddy power [url=https://luckyusaplay.com/#]best online casino bonus[/url] tips casino online
Hello There. I found your blog using msn. This is
a very well written article. I will make sure to
bookmark it and return to read more of your useful info. Thanks for the post.
I’ll certainly comeback.
My blog post; betxhub 247
Hi there, You have done a fantastic job. I’ll certainly digg it and personally suggest to my friends.
I’m sure they’ll be benefited from this site.
Feel free to surf to my web page :: JUDI ONLINE
You actually reported it terrifically.
agen judi casino osg777 online online gambling casino online casino uk best
Beneficial forum posts. Thanks a lot.
gta online casino car this week legit online casino green casino online
You said this fantastically!
online casino lastschrift [url=https://uscasinoguides.com/#]best online usa casino[/url] free casino online slot games
You actually said this very well.
online casino gratis startguthaben top online casinos bandar betting cbet casino online
What’s up all, here every one is sharing these kinds of experience, thus it’s nice to
read this website, and I used to visit this webpage every day.
Look at my web page … ro top serv
Superb tips. Appreciate it.
leovegas casino online [url=https://usagamblinghub.com/#]best usa online casino[/url] are online casinos legal in india
Wonderful posts. With thanks.
online casino agent hiring work from home no experience online casinos with no deposit bonus australian online casino $2 deposit
Beneficial knowledge. Kudos!
gratis geld online casino [url=https://usacasinomaster.com/#]casino games free online[/url] new mexico online casinos
Awesome info. Cheers!
parx online casino promo online casino welcome bonus dl juwa 777 online casino
Thanks! I like it.
10 online casino [url=https://luckyusaplay.com/#]no deposit online casino[/url] online casino pa real money
Excellent forum posts. Cheers.
gta online casino all access points best online casinos that payout where’s the diamond casino in gta v online
Terrific forum posts. Appreciate it.
usa online casino with ach deposit online live casino online casino slots odds
You actually said it well.
golden lady casino online [url=https://uscasinoguides.com/#]online casino games free[/url] white lion online casino
Nicely spoken of course. !
nuovi casino online legit online casinos fortune to go online casino
Hi, after reading this amazing article i am also glad to share my know-how here
with mates.
my blog: ELECTRONIC VISA
Incredible a lot of helpful knowledge!
usa friendly online casinos with free daily slots tournaments [url=https://usagamblinghub.com/#]casino games free online[/url] best online casino ios
Many thanks! Quite a lot of data!
casinos bonos bienvenida gratis sin depГіsito en mГ©xico online 2024 free play online casino new 2019 online casinos
Incredible a good deal of helpful knowledge!
vt online casino bonus [url=https://usacasinomaster.com/#]online casinos with no deposit bonus[/url] 21 grand casino online
You have made your point quite nicely!.
gta 5 online continue to scope out the casino online usa casinos online casino penny slots
What a information of un-ambiguity and preserveness of precious familiarity concerning unpredicted feelings.
Feel free to visit my web blog bollyflix
Lovely stuff, Thank you!
best online casino pragmatic play live dealer online casino a casino online
Superb posts. Thanks!
agen betting maxbet casino online [url=https://uscasinoguides.com/#]new online casino usa ok[/url] jack’s casino online review
Perfectly spoken genuinely! .
casino online che accettano american express [url=https://usaplayerscasino.com/#]deposit bonus casino online[/url] bandar judi osg777 casino online
Amazing knowledge. Thanks.
online casino blazing star online live casinos online casino vacancies
Incredible a good deal of great data!
do online casinos cheat blackjack [url=https://usagamblinghub.com/#]best online casinos that payout[/url] alle neuen online casinos
Thanks! Valuable information!
real money online casino no deposit free spins play casino online free best online casinos 2018
You explained it really well!
malaysia online casino website [url=https://usacasinomaster.com/#]casino online casino[/url] online casino rollover
Kudos, Terrific information!
online casino real money india [url=https://luckyusaplay.com/#]live online casino[/url] rtg us online casinos no deposit
You revealed that adequately.
online casino bonusi best online casino mentor gta online diamond casino heist the big con
You really make it seem really easy along with your presentation however I to find this matter to be
really something that I believe I would by no
means understand. It kind of feels too complex and extremely wide for me.
I am having a look forward to your subsequent publish,
I’ll attempt to get the hold of it!
Here is my web blog :: Free Link Shortener
You said it adequately..
casino night invitation online monkeytilt best online casino online casino juicy vegas $100 no deposit bonus codes
Whats up this is somewhat of off topic but I was wanting
to know if blogs use WYSIWYG editors or if you have to manually code with HTML.
I’m starting a blog soon but have no coding expertise so I wanted to get guidance from someone with experience.
Any help would be greatly appreciated!
Also visit my webpage :: URL shortener algorithm
You said it perfectly..
ignition casino online poker [url=https://usagamblinghub.com/#]best online casinos that payout[/url] no deposit bonus codes online casino
If some one needs expert view on the topic of running a blog then i
suggest him/her to visit this web site, Keep up the pleasant job.
Feel free to visit my site: אידוי קנאביס
Regards! Loads of facts!
river rock casino online [url=https://usacasinomaster.com/#]best online casinos[/url] best international online casino
You said it very well.!
pa mgm online casino us online casinos with rtg software casino sanremo online
You actually explained this well.
new online casino usa real money [url=https://luckyusaplay.com/#]casino games free online[/url] analisis casinos online colombia
You’ve made your point quite clearly.!
magic red online casino online casino free bonus crazy casino online
Regards, I like it!
2022 online casinos real money [url=https://uscasinoguides.com/#]legit online casino[/url] stake online casino location
You actually revealed that wonderfully!
ocean casino online gambling new online usa casinos simple casino online
You said it very well.
online casino in canada most trusted online casinos for usa players online casino ph
Many thanks, I like this.
free online casino win real money usa online casino free play no deposit ontario casinos online
You stated it well.
casino cyprus online [url=https://luckyusaplay.com/#]no deposit online casino[/url] online casinos portugal
buy generic cyproheptadine for sale – brand cyproheptadine 4mg tizanidine 2mg over the counter
Reliable advice. Thanks a lot.
online casino apps pa casino online no deposit online casino ireland no deposit
This is nicely said! .
casino online argentina dinero real [url=https://uscasinoguides.com/#]casino online usa[/url] real money online casino no deposit bonus usa
Thank you. Loads of stuff.
best online casinos india [url=https://usaplayerscasino.com/#]casino gambling online[/url] river sweepstakes online casino
Kudos, Ample content!
wild casino online real money newest online casinos usa steaks online casino
Valuable facts. Thanks a lot.
caesars online casino review [url=https://usagamblinghub.com/#]online casino with no deposit bonus[/url] online casino mastercard withdrawal
This is nicely said! .
ohio online casino gambling best online casino usa casino book of ra online
Many thanks. Lots of stuff!
south african online casinos with no deposit coupons [url=https://usacasinomaster.com/#]best online casinos[/url] online casino foxwoods
Many thanks! An abundance of postings.
how to win online casino jackpot [url=https://luckyusaplay.com/#]online casino top[/url] online casinos mit sofortГјberweisung
Perfectly expressed certainly! !
online casinos that accept echeck payments https://usagamblinghub.com/minnesota-casinos/ casino online bonos sin deposito
How can I import my blog to another gmail account?
https://disqus.com/by/axiomatiq/about/
Reliable forum posts. Regards!
online casino beginnen https://luckyusaplay.com/busr-review/ online real money casino free spins
Excellent write ups. Thank you!
online casinos no deposit bonus free spins https://uscasinoguides.com/betonline-review/ massachusetts online casinos
You actually stated this perfectly.
bally’s casino online gambling https://uscasinoguides.com/ohio-casinos/ eldorado online casino
Je n’était qu’un simple étudiant au baccalauréat.
my homepage; Gia
Awesome data, Thanks.
platinum play online casino free https://luckyusaplay.com/superbowl-betting/ mountaineer casino online
You actually reported that well.
agen sbc168 casino online https://usaplayerscasino.com/casino-games/ morongo casino play online
You reported it wonderfully!
online casino winning real money https://usacasinomaster.com/canadian-casinos/ royal vegas online casino app
Thank you! Loads of postings.
real money online casino nevada https://luckyusaplay.com/shazam-review/ rsweeps online casino 777 login
This is nicely expressed! .
san manuel casino online login https://usaplayerscasino.com/illinois-casinos/ best wv online casino
diclofenac price – order nimotop for sale purchase nimodipine sale
Amazing facts. Kudos.
casino atlantic city online https://usaplayerscasino.com/xbet-review/ online casino free bonus no deposit no wagering
Seriously many of valuable info.
online casino paypal deutschland https://luckyusaplay.com/crypto-casinos/ woo casino online
buy lioresal no prescription – order baclofen 10mg generic piroxicam price
Truly many of terrific material!
marina casino online https://usaplayerscasino.com/ethereum-casinos/ online casino med zimpler
Nicely put. Cheers.
golden nugget online casino new jersey https://luckyusaplay.com/florida-casinos/ top online casinos in south africa
You stated that perfectly!
bwin online casino https://uscasinoguides.com/washington-casinos/ agen joker123 casino online
You said that terrifically!
silversands online casino login https://uscasinoguides.com/credit-cards/ new online casinos canada
Nicely put. Appreciate it!
online casino gratis geld https://uscasinoguides.com/georgia-casinos/ nebraska online casino bonus
Thanks a lot, Lots of postings.
online casino education https://usacasinomaster.com/no-deposit-casinos/ kryptowährung online casino
Thanks. Plenty of write ups.
premier online casino https://uscasinoguides.com/poker-games/ online casino real money no minimum deposit
Fine data. Cheers!
gvc online casino https://usacasinomaster.com/virginia-casinos/ starlight online casino
It’s truly a great and useful piece of information. I’m happy that you shared this
helpful information with us. Please keep us
up to date like this. Thank you for sharing.
My page … get visibility with seo and markerting (Donte)
Awesome info. Thanks a lot.
online casino that accepts https://usagamblinghub.com/new-zealand-casinos/ casino online bonuses
Truly loads of helpful data.
online casinos that accept visa debit https://luckyusaplay.com/indiana-casinos/ online casino bonus ohne einzahlung 2022
Truly tons of excellent tips.
https://usagamblinghub.com/superbowl-betting/
What if i created a blog with the same name as other blog accidentally? is that legal?
https://offcourse.co/users/profile/john-leer
mestinon 60mg us – mestinon 60 mg ca buy azathioprine generic
buy rumalaya cheap – generic endep purchase endep generic
Thanks, Numerous postings!
https://uscasinoguides.com/xbet-review/
I am using the revolution code blue template for wordpress. I would like to change all fonts to Trebuchet MS. I have tried editing the stylesheet but no luck. Any suggestions?.
https://ralph.bakerlab.org/team_display.php?teamid=26243
How Can I Copyright Protect Stories and Articles on My Website?
https://ralph.bakerlab.org/team_display.php?teamid=26243
order voltaren 100mg generic – order aspirin 75mg for sale where to buy aspirin without a prescription
Very interesting information!Perfect just what I was looking for!
https://youtu.be/tzJ4u2-cdaQ
deflazacort oral – brimonidine sale alphagan online order
Great beat ! I wish to apprentice even as you amend your web site, how could i subscribe for a weblog website? The account aided me a appropriate deal. I have been tiny bit acquainted of this your broadcast offered vibrant clear idea
https://www.tinyhealth.com/blog/pregnancy-safe-medications
I think you have mentioned some very interesting points, thankyou for the post.
https://youtu.be/cuS-fXIYzj8
duphalac for sale online – betahistine 16mg drug betahistine 16 mg us
imusporin medication – buy cyclosporine generic buy colcrys no prescription
Utterly indited content material, Really enjoyed reading through.
https://youtu.be/wpchCZ_jguI
buy generic oxcarbazepine online – pirfenidone cost cheap levothroid pills
speman over the counter – finasteride price finasteride canada
you might have an awesome weblog right here! would you prefer to make some invite posts on my weblog?
https://youtu.be/3mAxNJ_g2_c
It’s a shame you don’t have a donate button! I’d definitely donate to this outstanding blog! I guess for now i’ll settle for book-marking and adding your RSS feed to my Google account. I look forward to brand new updates and will share this website with my Facebook group. Chat soon!
https://youtu.be/bqeueF7oPzE
Wow! Thank you! I continually needed to write on my blog something like that. Can I include a part of your post to my website?
https://youtu.be/yW59aXmGVfE
Attractive section of content. I just stumbled upon your site and in accession capital to say that I acquire in fact loved account your weblog posts. Anyway I’ll be subscribing in your feeds and even I achievement you get admission to constantly rapidly.
https://youtu.be/B16dCq5p6FI
Greetings! This is my first visit to your blog! We are a collection of volunteers and starting a new initiative in a community in the same niche. Your blog provided us beneficial information to work on. You have done a marvellous job!
https://rj-rajaslot88.com
he blog was how do i say it… relevant, finally something that helped me. Thanks
https://youtu.be/0Ei89v1YNYo
buy generic gasex for sale – buy generic gasex over the counter buy diabecon for sale
cheap lasuna tablets – order himcolin online cheap himcolin generic
order atorvastatin for sale – cost bystolic 5mg bystolic online order
Virtually all of whatever you mention happens to be astonishingly accurate and it makes me wonder the reason why I hadn’t looked at this with this light previously. This particular piece truly did switch the light on for me as far as this specific subject goes. However at this time there is 1 factor I am not really too comfy with and while I attempt to reconcile that with the actual core theme of your position, let me see what all the rest of your visitors have to point out.Nicely done.
https://youtu.be/2EAExRR5Du0
buy generic atenolol online – tenormin medication coreg without prescription
very nice post, i definitely love this web site, keep on it
https://youtu.be/htCNlttdDcs
I¦ll immediately grasp your rss as I can not in finding your e-mail subscription link or newsletter service. Do you’ve any? Kindly allow me understand so that I may subscribe. Thanks.
https://youtu.be/FgevmHDERA0
durex gel where to buy – order generic zovirax where to buy latanoprost without a prescription
zofran 4mg ca – buy tolterodine for sale ropinirole price
order generic cyclobenzaprine 15mg – primaquine over the counter buy generic enalapril for sale
Somebody essentially help to make seriously posts I would state. This is the very first time I frequented your web page and thus far? I amazed with the research you made to create this particular publish amazing. Wonderful job!
https://youtu.be/Iwo1I4DrVhM
Hiya, I am really glad I have found this information. Nowadays bloggers publish just about gossips and internet and this is actually irritating. A good web site with interesting content, that is what I need. Thanks for keeping this website, I will be visiting it. Do you do newsletters? Can not find it.
https://youtu.be/n3ZJVtprREU
order spironolactone 100mg without prescription – order dipyridamole generic naltrexone 50 mg drug
buy cyclophosphamide without prescription – cheap stavudine generic order trimetazidine without prescription
how to get norpace without a prescription – thorazine 50 mg us buy chlorpromazine 100mg generic
order divalproex for sale – purchase mefloquine pills oral topiramate 100mg
What is Lottery Defeater Software? Lottery Defeater Software is a plug-and-play Lottery Winning Software that is fully automated. Kenneth created the Lottery Defeater software. Every time someone plays the lottery, it increases their odds of winning by around 98.
https://youtu.be/Wb3pO9Kra0M
I have been surfing online greater than 3 hours lately, yet I by no means found any attention-grabbing article like yours. It¦s beautiful price sufficient for me. Personally, if all webmasters and bloggers made good content as you probably did, the web might be much more helpful than ever before.
https://youtu.be/VEXFdh00ODk
Hello my family member! I want to say that this post is amazing, nice written and include approximately all important infos. I¦d like to see extra posts like this .
https://www.axilusonline.com/hire-a-hacker-for-instagram/
I was just seeking this information for some time. After six hours of continuous Googleing, finally I got it in your web site. I wonder what is the lack of Google strategy that do not rank this kind of informative web sites in top of the list. Generally the top websites are full of garbage.
https://youtu.be/RZj5HVtZ4pQ
piroxicam brand – buy exelon without a prescription rivastigmine brand
purchase enalapril online – doxazosin 2mg pill purchase latanoprost generic
buy dramamine 50 mg online – purchase dramamine online order actonel pills
ProvaDent: What Is It? ProvaDent is a natural tooth health supplement by Adam Naturals.
https://youtu.be/bai1s45JdnY
cost fulvicin – fulvicin 250 mg drug gemfibrozil 300 mg generic
buy forxiga generic – how to buy dapagliflozin brand precose 50mg
Good write-up, I am normal visitor of one?¦s web site, maintain up the excellent operate, and It is going to be a regular visitor for a long time.
https://www.profi-poolwelt.de/300-x-125-cm-aufstellpool-set.html
Its like you read my mind! You seem to know a lot about this, like you wrote the book in it or something. I think that you can do with a few pics to drive the message home a bit, but other than that, this is wonderful blog. An excellent read. I’ll definitely be back.
https://pool.net/achtformbecken-8-55-x-5-00-x-1-50-m.html
buy eukroma online – desogestrel 0.075mg cheap dydrogesterone brand
generic cotrimoxazole 960mg – buy generic bactrim online order tobrex 5mg drops
order bisacodyl 5mg generic – buy liv52 online cheap liv52 20mg brand
fludrocortisone helmet – esomeprazole pills set prevacid pills research
clarithromycin pills fleet – asacol fearful cytotec practical
This is very interesting, You are a very skilled blogger. I have joined your rss feed and look forward to seeking more of your fantastic post. Also, I have shared your site in my social networks!
http://www.tlovertonet.com/
promethazine ponder – promethazine somewhat promethazine lack
ascorbic acid sink – ascorbic acid item ascorbic acid represent
If you want to take a good deal from this piece of
writing then you have to apply these methods to your won website.
My blog – nordvpn special coupon code 2024
Howdy! I know this is kind of off topic but I was
wondering which blog platform are you using for this site?
I’m getting fed up of WordPress because I’ve had issues with hackers and I’m looking at options for another platform.
I would be great if you could point me in the direction of a
good platform.
Also visit my homepage eharmony special coupon code 2024
dapoxetine struggle – dapoxetine task priligy chuckle
loratadine overhead – claritin pills mouth claritin pills bridge
valacyclovir salt – valacyclovir pills half valacyclovir good
pills for treat prostatitis shin – prostatitis treatment recover prostatitis pills back
treatment for uti frighten – uti treatment read uti medication specter
acne medication ticket – acne treatment data acne medication side
cenforce secure – kamagra mankind brand viagra online wander
Pretty! This has been an incredibly wonderful article. Many thanks
for supplying these details.
My site facebook vs eharmony to find love online
cialis soft tabs online fact – cialis super active struggle1 viagra oral jelly advantage
brand cialis slay – forzest stare penisole speed
brand cialis greater – alprostadil roll penisole draught
viagra professional online shadow – eriacta release levitra oral jelly underground
simvastatin communication – lopid god lipitor accomplish
rosuvastatin west – ezetimibe buy poison caduet buy attach
nitroglycerin order online – buy generic indapamide online generic valsartan
buy metoprolol 50mg generic – buy generic lopressor adalat cost
hydrochlorothiazide 25 mg pill – buy generic norvasc zebeta price
digoxin 250mg usa – calan without prescription furosemide 100mg pills
famvir pills – order acyclovir 400mg valcivir for sale online
buy nizoral medication – purchase lotrisone cream sporanox 100mg ca
order semaglutide without prescription – glucovance uk desmopressin cost
buy generic lamisil online – buy generic terbinafine for sale buy griseofulvin sale
micronase 2.5mg uk – brand glucotrol where to buy dapagliflozin without a prescription
hzPJBYMtUZuc
order desloratadine 5mg – desloratadine for sale online buy albuterol generic
Hi,
I just visited bioengx.com and wondered if you’d ever thought about having an engaging video to explain what you do?
I can show you some previous videos we’ve done if you want me to send some over. Let me know if you’re interested in seeing samples of our previous work.
Regards,
Georgina
Unsubscribe: https://removeme.click/ev/unsubscribe.php?d=bioengx.com
purchase albuterol inhalator sale – order fluticasone online theo-24 Cr 400 mg drug
stromectol ivermectin – cefaclor pill where can i buy cefaclor
cheap azithromycin 250mg – buy tinidazole 500mg sale ciplox price
buy cheap clindamycin – order suprax 100mg buy chloromycetin without prescription
You actually make it appear really easy along with your presentation however I to find this topic to be actually
one thing that I feel I’d by no means understand.
It seems too complicated and extremely large for me. I’m looking forward for your subsequent post, I’ll try
to get the hold of it!
Feel free to surf to my web-site vpn coupon code 2024
Hi there! Would you mind if I share your blog with my zynga group?
There’s a lot of people that I think would really enjoy your content.
Please let me know. Thanks
Feel free to surf to my web site; vpn special code
amoxil us – purchase ceftin for sale order cipro 500mg generic
order augmentin 375mg without prescription – zyvox ca cipro price
atarax cost – generic amitriptyline 25mg endep ca
buy generic anafranil 50mg – order aripiprazole 20mg pills purchase sinequan sale
order clozaril pill – order frumil 5mg sale famotidine buy online
zidovudine medication – lamivudine online buy buy zyloprim sale
order metformin without prescription – order baycip lincomycin drug
lasix 100mg usa – atacand 16mg tablet captopril for sale
ampicillin online buy amoxil without a prescription buy amoxil for sale
flagyl ca – buy cefaclor 500mg zithromax for sale online
ivermectin 6 mg otc – sumycin us buy tetracycline 500mg without prescription
valacyclovir 1000mg drug – nemasole over the counter order acyclovir online cheap
ciprofloxacin 500 mg oral – buy doxycycline tablets order erythromycin 500mg pills
brand flagyl – order flagyl 200mg pills buy azithromycin 250mg sale
Hi there,
We run a Youtube, Instagram and Twitter/X service, where we can increase your subscribers/followers.
This is all done manually and so all subscribers and followers are real people.
Our prices start from just $60/month
If you are interested, please let us know, which of your social media profiles are you wishing to grow?
If you are not interested, no problem, just ignore and delete this email.
Kind Regards,
Sarah
baycip price – cephalexin buy online buy amoxiclav generic
buy cipro 1000mg generic – myambutol 600mg sale order augmentin 375mg for sale
lipitor 10mg canada buy atorvastatin 80mg pill order atorvastatin 20mg pill
VlrkbLCeda
创建relax_flag文件,文件内容如下:
-use_input_sc
-constrain_relax_to_start_coords
-ignore_unrecognized_res
-nstruct 20
-relax:coord_constrain_sidechains
-relax:cartesian-score:weights ref2015_cart # 务必设为ref2015_cart
-relax:min_type lbfgs_armijo_nonmonotone # 能量最小化的算法
-relax:script cart2.script
should be
-use_input_sc
-constrain_relax_to_start_coords
-ignore_unrecognized_res
-nstruct 20
-relax:coord_constrain_sidechains
-relax:cartesian
-score:weights ref2015_cart # 务必设为ref2015_cart
-relax:min_type lbfgs_armijo_nonmonotone # 能量最小化的算法
-relax:script cart2.script
Hi there,
We run a YouTube growth service, which increases your number of subscribers both safely and practically.
– We guarantee to gain you 700-1500 new subscribers per month.
– People subscribe because they are interested in your videos/channel, increasing video likes, comments and interaction.
– All actions are made manually by our team. We do not use any ‘bots’.
The price is just $60 (USD) per month, and we can start immediately.
If you’d like to see some of our previous work, let me know, and we can discuss it further.
Kind Regards,
Emily